The CRISPR/Cas9 genome editor is rapidly changing biomedical research, but the new technology is not yet clear. This technique can inadvertently produce excessive or unwanted changes in the genome and produce non-target gene mutations that limit safety and efficacy in therapeutic applications.
Now, a new study published by Cell, researchers at the Massachusetts Medical School and the University of Toronto, discovered the first known "off switch" for CRISPR/Cas9 activity, providing greater controllability for editors.

Erik J. Sontheimer, PhD, professor in the RNA Therapeutics Institute at UMass Medical School Credit: University of Massachusetts Medical School
Image source: Medical Press
Dr. Erik J. Sontheimer, Professor of RNA Therapy at the Massachusetts Medical School, Professor Alan Davidson of Molecular Genetics and Dr. Karen Maxwell, Assistant Professor of Biochemistry at the University of Toronto, identified three naturally occurring proteins that inhibit the Cas9 enzyme. These proteins, called anti-CRISPR, have the ability to block DNA cleavage by Cas9 nucleases.
Dr. Sontheimer said, "CRISPR / Cas9 is very useful because it introduces specific chromosome breaks that can be used to create genome edits, but because chromosome breaks can be harmful and may last too long. There is no reliable way to shut down. Cas9, once it has been edited at the cell, if it can be closed after the correct editing is completed, then the problem is solved. Here we report the first known inhibitor of Cas9 natural activity."
Dr. Davidson said, "CRISPR is very powerful, but we must be able to close its editor. This is a very basic toolbox that will give researchers more confidence in using gene editing."
CRISPR / Cas9 is an adaptive immune defense formed by bacteria and archaea during long-term evolution and can be used against invading viruses and foreign DNA. It consists of two parts: the molecular knife (Cas9), which effectively cleaves DNA, but is latched in its natural state; the RNA-directed complex unlocks Cas9 when a sequence of complementary bases is found, thus determining precise cutting Site. These guide RNAs are generated by CRISPR "aggregated regularly spaced short palindromic repeats" or CRISPR arrays containing residues of past virally infected genomes. Targeting cleavage and inactivation of these viruses by Cas9 nucleases, CRISPR/Cas9 provides adaptive immune defenses for bacterial cells.
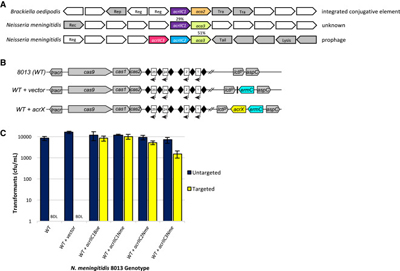
Identification and Validation of Type II-C Anti-CRISPRs
Image source: Cell
Lab Microscope,Binocular Biological Lab Microscope,Binoculars Lab Optical Microscope,Laboratory Biological Optical Microscope
NINGBO VANCO INSTRUMENT CO.,LTD , https://www.vancoscope.com