Over the past 20 years, the development of high-throughput technologies has supported large-scale genomic, transcriptome, and proteomic quantitative analysis. These data were used to analyze the quantitative relationship between transcriptome and proteome under different systems and conditions. These studies sometimes lead to conflicting conclusions, especially as to what extent the amount of mRNA controls the amount of protein.
A core issue in basic life science research and transformational life science research is how genomic information determines phenotype through its own expression. The central rule closely links the three molecules of DNA, RNA and protein. The relationship between the amount of transcripts derived from the same locus and the amount of protein is not simple. In addition to the regulation of the amount of transcripts, there are many other important regulatory pathways. These pathways include: (1) translation rate: translation rate is significantly affected by mRNA sequences, such as the upstream open reading frame, the internal ribosome binding site; (2) the regulation of translational rate: the regulation of protein binding to transcripts Regulatory elements, non-coding RNA binding, transcript binding ability to ribosomes; (3) protein half-life: degradation of ubiquitination pathway or autophagy independent of transcripts; (4) protein synthesis Delay: Protein synthesis takes time, so transcriptional changes require a short delay to affect protein levels; (5) Protein transport: Proteins are spatially transported out of where they are synthesized. Therefore, it is not advisable to directly compare protein abundance with mRNA abundance.
There are two different forms of protein to mRNA comparison. One is (Figure 1A) comparing the amount of protein and mRNA derived from the same gene at different individuals, conditions, or time points. This is done to investigate the extent to which changes in mRNA levels affect changes in protein levels. . The other (Figure 1B) compares the amount of a number of different proteins and their corresponding mRNAs in order to investigate the extent to which protein levels can reflect mRNA differences.
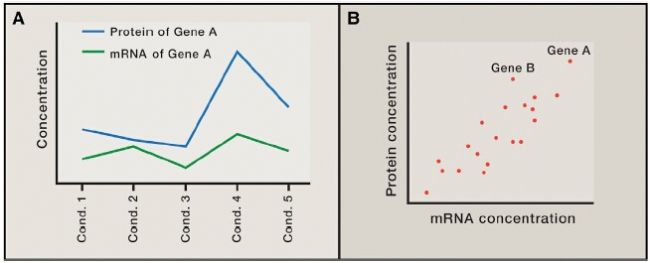
Figure 1. Different forms of protein and mRNA are compared with each other.
Y Liu, A Beyer, R Aebersold. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell.
Advances in technology have deepened our understanding of the relationship between mRNA and protein levels. Genome-level research requires high-precision, high-sensitivity, and high-accuracy techniques (Figure 2).
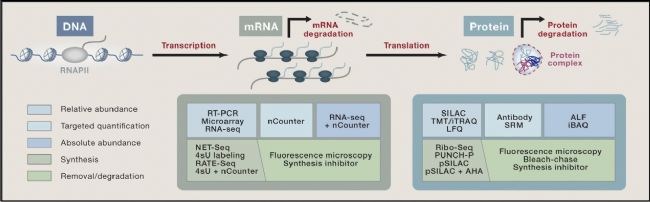
Figure 2. A series of mechanisms for controlling gene expression and corresponding quantitative methods
Y Liu, A Beyer, R Aebersold. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell.
Mass spectrometry has become an important method for studying protein characterization and quantification over the past 20 years. As a comprehensive tool, mass spectrometry supports large-scale relative and absolute quantification without the need for antibodies (Table 1).
Table 1 Mass Spectrometry Technology Summary and Representative Literature for Studying the Relationship between mRNA and Protein
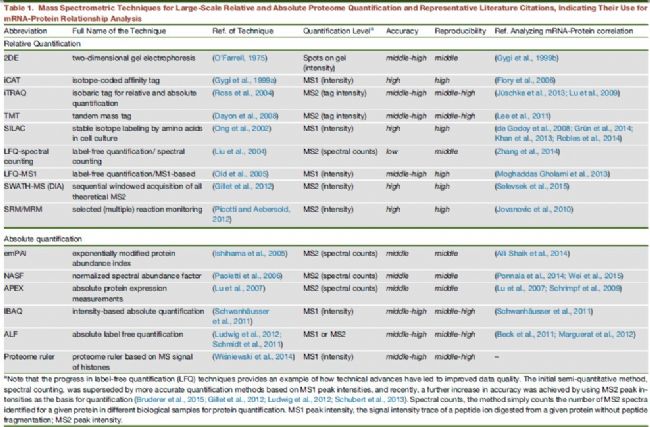
Y Liu, A Beyer, R Aebersold. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell.
At steady state, the level of mRNA determines the level of protein, and protein synthesis is delayed (Figures 3A and 3B). It is difficult to strictly define the "stable state". Let us think that the cells we usually collect during the omics experiment, if the protein or mRNA level in a certain period of time (usually a few hours) remains relatively constant, it is regarded as "Stable state."
Post-transcriptional regulation is very important during a brief adaptation period. If you change the living environment and conditions of the cells, the cells need a short adaptation period. At this time, if only changing the transcription level is too slow, a post-transcriptional regulation mechanism is required. Increasing the translation rate of existing transcripts can rapidly synthesize new desired proteins while target-degrading proteins, such as the ubiquitin pathway, to accelerate the elimination of unwanted proteins and even toxic proteins (Figures 3B–3D). The mechanism of on-demand expression ensures that cells respond to the synthesis of the desired protein rapidly during signal stimulation rather than constitutively expressing the protein (Figures 3B). On the other hand, it is well known that post-translational modifications (which can also be considered as a post-transcriptional regulation) are also important during the brief adaptation period.
The synthesis and degradation of intracellular proteins are balanced due to energy and concentration limitations. Protein synthesis is limited in many ways. On the one hand is energy and nutrition; on the other hand is the regulation mechanism of catalytic protein synthesis. Synthetic proteins cost much more than synthetic mRNAs, so cells choose the most economical way to solve the problem of protein synthesis. Despite various limitations, cells strive to keep proteins at a constant level, whether steady or dynamic. This physical limitation can be used to optimize protein diffusion rates, protein transport between cells, and biochemical reaction rates (Figures 3E–3G). Within a cell, the copy number of the protein is much greater than the copy number of the mRNA. A study showed that when the fission yeast transitioned to equilibrium during the proliferative phase, the copy number of mRNA was reduced by nearly 70%, while the copy number of protein was only reduced by 9.5%.
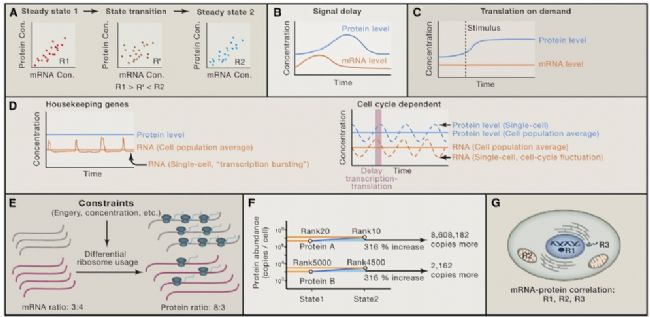
The importance of the dynamic relationship between Figures 3 mRNA and protein
Y Liu, A Beyer, R Aebersold. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell.
The synthesis of proteins requires cells to allocate resources, and this distribution begins with transcriptional regulation. The cost of producing a protein by a cell is primarily determined by the precursor of the protein, the composition of the protein (such as molecular weight), and the rate of protein synthesis. Studies have shown that in bacterial metabolic pathways, synthetic high-cost enzymes are usually regulated by multiple transcriptional rates; in contrast, synthetic low-cost enzymes are only controlled by several key reactions, which are usually constitutively expressed. Therefore, synthetic high-cost enzymes are tightly regulated to avoid "wasting", and low-cost enzymes can be expressed even if they are not needed. Transcriptional regulation reflects the balance between minimally costly synthetic proteins and maximizing function. Studies have shown that in a particular cell cycle, if you want to activate or inactivate a protein complex, you don't have to synthesize or degrade each member of the complex. It is enough to adjust some key members. Using this economic principle, cells regulate protein complexes with minimal cost. However, in multicellular organisms, this optimized mechanism may be different.
Cells are allocated resources through the use of ribosomes. Cells have to be flexible in using translational machinery primitives such as translation factors, tRNA, aminoacyl tRNA synthetase and ribosomes. The main limitation of yeast protein translation may be competition for free ribosomes rather than competition for aminoacyl tRNA.
The regulation of the absolute and relative amounts of high abundance proteins. Up-regulation of high-abundance proteins is much more expensive than up-regulation of multiples of low-abundance proteins, requiring more copy number. High abundance proteins are typically produced by high mRNA absolute levels and efficient translation. In other words, the amount of mRNA and the rate of translation are positively correlated with protein abundance. However, the rate of production of highly expressed proteins appears to be saturated, suggesting that overall translation has become a limitation of most high-abundance proteins. This limitation is most likely due to ribosome saturation on the mRNA. Therefore, those high-abundance proteins usually have the lowest rate of protein change and usually perform the function of housekeeping proteins.
On the other hand, recent studies have shown that differences in protein synthesis rates are major factors in determining protein copy number differences (Figures 3F). This situation suggests that changes in mRNA can determine changes in the relative amount of protein, and post-transcriptional regulation leads to more changes in the absolute level of the protein. Thus, although post-translational regulation may significantly affect protein copy number, it may not fundamentally affect relative changes between proteins.
Different spaces have different relationships between proteins and mRNA. Different spaces, such as cell populations and tissues and single cells, single cells and subcellular units, also significantly determine the extent to which the amount of protein reflects the amount of transcript. If the cell type is not distinguished in the tissue, it will lead to a wrong assessment of the relationship between protein and mRNA. The correlation between the single intracellular protein and the corresponding mRNA is reduced, and the correlation between the whole cell level and the subcellular level is also different.
In summary, the level of protein in steady state is largely determined by the amount of transcript; in dynamic stages, such as cell differentiation or stress, the post-transcriptional process may lead to more changes. In many scenarios, protein levels cannot be adequately predicted based solely on the level of transcription. Therefore, to explain the relationship between genotype and phenotype, high-quality multi-level gene expression data is essential for a complete understanding of biological processes. The development of proteomics has made it possible for this, and iTRAQ, SILAC, SWATH and other technologies have been widely used in the research of agriculture, industry, medicine and other industries.
references
Y Liu, A Beyer, R Aebersold. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell, 2016, 165(3): 535-550
Fuvision 4g Security HD 360 Degree Ip Dmart Wireless Home Security Camera 360 WIFI Audio Night Vision Cloud CCTV Camera Fuvision 4g Security HD 360 Degree Ip Dmart Wireless Home Security Camera 360 WIFI Audio Night Vision Cloud CCTV Camera
1. Indoor devices: such as speakers, sockets, chargers, lamps, picture frames. The micro camera is ingeniously placed in these objects, so as to monitor the home environment and protect the personal and property safety in the home.
2. Personal jewelry shape: watch, glasses, tie, lighter, badge. The camera is made into the shape of an ornament, which can facilitate secret visits and recording.
3. Form of articles for daily use: tissue box, beverage can, car key, light switch, row plug, alarm clock, flashlight. The camera of this shape can be placed at the position for monitoring at will.
Distinguish from functions:
1. Common linear type
2. Wireless transmitting type
3. Infrared night vision type
4. Ultra long distance transmission type
China 4G CCTV Camera, low power consumption supplier & manufacturer, offer low price, high quality 4g Outdoor Wireless Camera,Solar Camera, etc.
4G CCTV Camera, low power consumption,4g Outdoor Wireless Camera,Solar Camera 4G CCTV,Solar Powered CCTV Outdoor Camera
Shenzhen Fuvision Electronics Co., Ltd. , https://www.outdoorsolarcamera.com