--- Jia Wei Waters Technology (Shanghai) Co., Ltd. Experimental Center
The (primary) parent ion and (secondary) fragment ion data of the unknown are the information necessary for mass spectrometry. In addition to the DDA tandem mass spectrometry acquisition method, Waters Mass Spectrometry offers a unique Total Information Tandem Mass Spectrometry (MS E ) technology. So how does MS E technology get serial information and optimize and maximize information collection?
What kind of information does full information tandem mass spectrometry (MS E ) provide?
1. Qualitative and quantitative analysis of unknown analytes is done in the same analysis.
2. Simultaneously obtain high resolution, high quality and accurate data of parent ions and fragment ions.
3. MS E is universally applicable to a variety of unknowns analysis, and the method setup is very simple.
4. Give full play to the superior performance of UPLC-MS LC/MS.
What is Full Information Tandem Mass Spectrometry (MS E )?
1. MS E is a tandem mass spectrometry method that simultaneously obtains highly accurate precursor and fragment ion information in a liquid chromatography analysis.
2. MS E consists of two scans: “no collision energy†and “high collision energyâ€, recording parent ion and fragment information separately.
3. MS E performs the parent-child ion association by the same chromatographic behavior of the parent ion and its fragment ions.
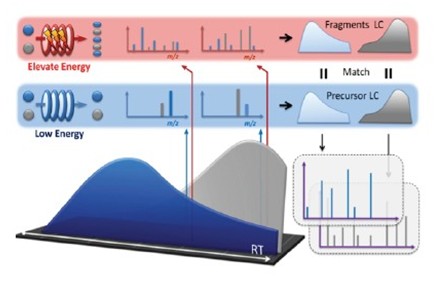
What are the characteristics of full information tandem mass spectrometry (MS E )?
1. Comprehensive: All ion information is recorded, more accurate and more accurate.
2. Accuracy: All parent ion and fragment ion information are high-precision, high-resolution mass spectrometry data.
3. Simple: The method setting only needs three parameters: mass range, acquisition time and collision energy.
4. Flexibility: Collision energy is a linear increase, so different analytes can be fragmented at their optimal collision energy.
What are the advantages of MS E compared to conventional DDA tandem mass spectrometry?
Data-dependent tandem mass spectrometry (DDA. Data Dependent Acquisition) is to collect the corresponding fragment ions by selecting specific precursor ions into the collision cell. MS E does not select a specific parent ion for individual fragmentation, but simultaneously collects fragment ions of all parent ions. In this way, MS E avoids the problem of incomplete information collection due to the limitation of DDA acquisition rate. In addition, MS E's uniform high-frequency data acquisition mode provides a “perfect†chromatogram for each ion for accurate quantification. In contrast, DDA often has defects in the chromatographic peak due to the contingency of the acquisition, which affects the quantitative accuracy.

Why is MS E and UPLC the best partner?
UPLC ® in chromatographic resolution (selectivity), peak height (sensitivity) and run time (velocity) with respect HPLC are more qualitative leap. However, UPLC's short and slender peaks also place higher demands on mass spectrometry. On the one hand, the MS E mass spectrometry method subtly solves the limitation of the DDA acquisition frequency; on the other hand, UPLC also provides a solid foundation for the MS E method to achieve high and accurate maternal ion assignment.

MS E technology has been widely used in biopharmaceutical analysis, proteomics, metabolite identification, metabolomics, lipidomics, impurity identification, forensic toxicology, environmental analysis, food testing, chemical material analysis and other fields. application.
references
(1) Bateman, Carruthers, Hoyes, Jones, Langridge, Millar, Vissers; A novel precursor ion discovery method on a hybrid quadrupoleorthogonal acceleration time-of-flight (Q-TOF) mass spectrometer for studying protein phosphorylation, J. Am. Soc Mass Spectrom., 2002; 13, 792-803.
(2) Silva, Denny, Dorschel, Gorenstein, Kass, Li, McKenna, Nold, Ric hardson, Young, Geromanos; Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005 Apr 1;77(7):2187- 200.
(3) Blackburn K, Mbeunkui F, Mitra SK, Mentzel T, Goshe MB. Improving protein and proteome coverage through data-independent multiplexed peptide fragmentation. J. Proteome Res. 2010 Jul 2;9(7):3621-37.
(4) C ha kra borty AB, Berger SJ, Gebler JC. Use of an integrated MS-MS MS/MS data acquisition strategy for high coverage plant peptide studies. Rapid Commun. Mass Spectrom. 2007;21(5):730-44 .
(5) Tiller PR, Yu S, Castro-Perez J, Fillgrove KL, Baillie TA. Hight hroughput, accurate mass liquid c hromatography/tandem mass spectrometry on a quadrupole time-of-flight system as a 'first-line'approach for Metabolite identification studies. Rapid Commun. Mass Spectrom. 2008 Apr;22(7):1053-61.
(6) Simplified approac hes to impurity identification using accurate mass UPLC/MS; Waters Application Note, http:// 03850en.pdf
(7) T he utility of MSE for toxicological screening; Waters Technology Brief, http://
(8) A case of pesticide poisoning: T he use of a broad-scope Tof screening approach in wildlife protection; Waters Application Note, http://
(9) Addressing c hemical diversity and expanding analytical capabilities with APGC; Waters White Paper, http://
(10) McEwen, McKay; A combination atmospheric pressure LC/MS: GC/MS ion source: Advantages of dual AP-LC/MS: GC/MS instrumentation, J. Am. Soc. Mass Spectrom., 2007; 16, 1730 -1738.
Fresh Ginger,Ground Ginger Root,Minced Fresh Ginger,Minced Ginger Root
Yirun Agricultural Cooperative , https://www.yiruncn.com